FQ Group
Chemical Biology and Cell Biology
Chemical biology and cell biology
Research
Research
Research
The chromosome is mainly composed of nucleosomes formed by DNA entangled with histones, which are combined with a series of modification-interacting proteins, transcription factors, chromatin remodeling factors to form highly concentrated aggregates. Transposons, also known as 'jumping genes', can self-replicate and transfer to other random gene loci. They not only provide abundant genetic materials for biological evolution, but also play a key role in the adaptive evolution of species. They are one of the driving forces of genome evolution and an important source of biodiversity and complexity.
We are committed to spatio-temporal analysis of chromatin spatial proteome, and comprehensively apply interdisciplinary technology to the frontier research of decoding the mystery of life. We hope to provide a molecular theoretical basis for the screening and evolution of DNA fragments in the process of gene transposition, and the regulation of gene expression in time and space.
The chromosome is mainly composed of nucleosomes formed by DNA entangled with histones, which are combined with a series of modification-interacting proteins, transcription factors, chromatin remodeling factors to form highly concentrated aggregates. Transposons, also known as 'jumping genes', can self-replicate and transfer to other random gene loci. They not only provide abundant genetic materials for biological evolution, but also play a key role in the adaptive evolution of species. They are one of the driving forces of genome evolution and an important source of biodiversity and complexity.
We are committed to spatio-temporal analysis of chromatin spatial proteome, and comprehensively apply interdisciplinary technology to the frontier research of decoding the mystery of life. We hope to provide a molecular theoretical basis for the screening and evolution of DNA fragments in the process of gene transposition, and the regulation of gene expression in time and space.
The chromosome is mainly composed of nucleosomes formed by DNA entangled with histones, which are combined with a series of modification-interacting proteins, transcription factors, chromatin remodeling factors to form highly concentrated aggregates. Transposons, also known as 'jumping genes', can self-replicate and transfer to other random gene loci. They not only provide abundant genetic materials for biological evolution, but also play a key role in the adaptive evolution of species. They are one of the driving forces of genome evolution and an important source of biodiversity and complexity.
We are committed to spatio-temporal analysis of chromatin spatial proteome, and comprehensively apply interdisciplinary technology to the frontier research of decoding the mystery of life. We hope to provide a molecular theoretical basis for the screening and evolution of DNA fragments in the process of gene transposition, and the regulation of gene expression in time and space.
The SiTomics technology (Cell, 2023, 186(5), 1066-1085) jointly developed by me and my collaborators decodes the distribution of specific interacting protein bindings mediated by different histone modifications at the genomic level. At the same time, inspired by the energy field principle of transcription factor binding DNA published by Stephen R. Quake (Science, 2007, 315, 233-237), the concept of chromatin-associated proteome was proposed, which successfully extended the space group to the field of chromosomes, that is, specific binding proteins are distributed in different genomic locations, and this concept have been recognized by peers.
The SiTomics technology (Cell, 2023, 186(5), 1066-1085) jointly developed by me and my collaborators decodes the distribution of specific interacting protein bindings mediated by different histone modifications at the genomic level. At the same time, inspired by the energy field principle of transcription factor binding DNA published by Stephen R. Quake(Science, 2007, 315, 233-237), the concept of chromatin-associated proteome was proposed, which successfully extended the space group to the field of chromosomes, that is, specific binding proteins are distributed in different genomic locations,and this concept have been recognized by peers.
The SiTomics technology (Cell, 2023, 186(5), 1066-1085) jointly developed by me and my collaborators decodes the distribution of specific interacting protein bindings mediated by different histone modifications at the genomic level. At the same time, inspired by the energy field principle of transcription factor binding DNA published by Stephen R. Quake (Science, 2007, 315, 233-237), the concept of chromatin-associated proteome was proposed, which successfully extended the space group to the field of chromosomes, that is, specific binding proteins are distributed in different genomic locations, and this concept have been recognized by peers.
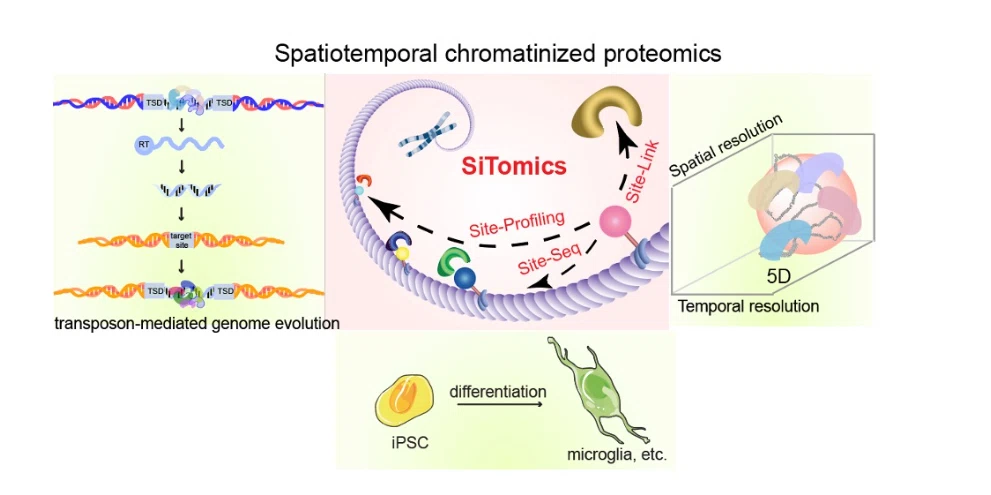
Based on the applicant 's previous research foundation and technical advantages, our research group will continue to promote the research direction of spatial proteomics and contribute to the vigorous development of the International Big Science Program for Human Proteome Navigation ('π-HuB Program') led and launched by Chinese scientists.
1.The development of chemical biology and molecular biology tools to analyze the dynamic changes of chromosome binding proteins in time and space provides a reference value for a comprehensive understanding of basic biological problems such as gene evolution and cell fate determination, and also provides a solid theoretical basis for the realization of time and space controllable gene expression.
2.Expanding chemical biology tools, accurately study the effect of site-specific protein post-translational modification on protein function, discovering and clarifyIng the dynamic characteristics of protein post-translational modification, revealing its biological effects and regulatory mechanisms, and obtain new drug targets and corresponding intervention small molecules for dynamic modification, in order to achieve targeted chemical intervention of protein post-translational modification, accelerate the transformation from basic research to drug development, and provide basic and forward-looking scientific and technological reserves for understanding the inherent laws of life system regulation, diagnosis and prevention of major diseases.
3.The iPSC was used as the research system to study the effect of dynamic modification of biological macromolecules on cell differentiation and the relationship between metabolism and cell fate.
Based on the applicant 's previous research foundation and technical advantages, our research group will continue to promote the research direction of spatial proteomics and contribute to the vigorous development of the International Big Science Program for Human Proteome Navigation ('π-HuB Program') led and launched by Chinese scientists.
1.The development of chemical biology and molecular biology tools to analyze the dynamic changes of chromosome binding proteins in time and space provides a reference value for a comprehensive understanding of basic biological problems such as gene evolution and cell fate determination, and also provides a solid theoretical basis for the realization of time and space controllable gene expression.
2.Expanding chemical biology tools, accurately study the effect of site-specific protein post-translational modification on protein function, discovering and clarifyIng the dynamic characteristics of protein post-translational modification, revealing its biological effects and regulatory mechanisms, and obtain new drug targets and corresponding intervention small molecules for dynamic modification, in order to achieve targeted chemical intervention of protein post-translational modification, accelerate the transformation from basic research to drug development, and provide basic and forward-looking scientific and technological reserves for understanding the inherent laws of life system regulation, diagnosis and prevention of major diseases.
3.The iPSC was used as the research system to study the effect of dynamic modification of biological macromolecules on cell differentiation and the relationship between metabolism and cell fate.
Based on the applicant 's previous research foundation and technical advantages, our research group will continue to promote the research direction of spatial proteomics and contribute to the vigorous development of the International Big Science Program for Human Proteome Navigation ('π-HuB Program') led and launched by Chinese scientists.
1.The development of chemical biology and molecular biology tools to analyze the dynamic changes of chromosome binding proteins in time and space provides a reference value for a comprehensive understanding of basic biological problems such as gene evolution and cell fate determination, and also provides a solid theoretical basis for the realization of time and space controllable gene expression.
2.Expanding chemical biology tools, accurately study the effect of site-specific protein post-translational modification on protein function, discovering and clarifyIng the dynamic characteristics of protein post-translational modification, revealing its biological effects and regulatory mechanisms, and obtain new drug targets and corresponding intervention small molecules for dynamic modification, in order to achieve targeted chemical intervention of protein post-translational modification, accelerate the transformation from basic research to drug development, and provide basic and forward-looking scientific and technological reserves for understanding the inherent laws of life system regulation, diagnosis and prevention of major diseases.
3.The iPSC was used as the research system to study the effect of dynamic modification of biological macromolecules on cell differentiation and the relationship between metabolism and cell fate.
.png)